
Data from Google Scholar (update on November, 2024) Web Link
M. Luo, Y. Yin, Z. Zhou, H. Zhang, X. Chen, H. Wang, and Z.-J. Zhu*, A Mass Spectrum-oriented Computational Method for Ion Mobility-resolved Untargeted Metabolomics, Nature Communications, 2023, 14: 1813. Web Link
H. Wang†, H. Jia†, Y. Gao, H. Zhang, J. Fan, L. Zhang, F. Ren, Y. Yin, Y. Cai*, J. Zhu*, and Z.-J. Zhu*, Serum Metabolic Traits Reveal Therapeutic Toxicities and Responses of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer, Nature Communications, 2022, 13: 7802. Web Link
Z. Zhou†, M. Luo†, H. Zhang, Y. Yin, Y. Cai, and Z.-J. Zhu* , Metabolite Annotation from Knowns to Unknowns through Knowledge-guided Multi-layer Metabolic Networking, Nature Communications, 2022, 13: 6656. Web Link
R. Wang, Y. Yin, J. Li, H. Wang, W. Lv, Y. Gao, T. Wang, Y. Zhong, Z. Zhou, Y. Cai, X. Su, N. Liu*, and Z.-J. Zhu*, Global Stable-isotope Tracing Metabolomics Reveals System-wide Metabolic Alternations in Aging Drosophila, Nature Communications, 2022, 13: 3518. Web Link
X. Mei†, Y. Guo†, Z. Xie†, Y. Zhong, X. Wu, D. Xu, Y. Li, N. Liu, and Z.-J. Zhu*, RIPK1 Regulates Starvation Resistance by Modulating Aspartate Catabolism, Nature Communications, 2021, 12: 6144. Web Link
T. Li, Y. Yin, Z. Zhou, J. Qiu, W. Liu, X. Zhang, K. He, Y. Cai, and Z.-J. Zhu*, Ion Mobility-based Sterolomics Reveals Spatially and Temporally Distinctive Sterol Lipids in the Mouse Brain, Nature Communications, 2021, 12: 4343. Web Link
Z. Zhou, M. Luo, X. Chen, Y. Yin, X. Xiong, R. Wang, and Z.-J. Zhu*, Ion Mobility Collision Cross-Section Atlas for Known and Unknown Metabolite Annotation in Untargeted Metabolomics,Nature Communications, 2020, 11: 4334. Web Link
X. Shen, R. Wang, X. Xiong, Y. Yin, Y. Cai, Z. Ma, N. Liu, and Z.-J. Zhu*,Metabolic Reaction Network-based Recursive Metabolite Annotation for Untargeted Metabolomics, Nature Communications, 2019, 10: 1516. Web Link
Citrulline regulates macrophage metabolism and inflammation to counter aging in mice
Z. Xie, M. Lin, B. Xing, H. Wang, H. Zhang, Z. Cai, X. Mei*, and Z.-J. Zhu*
Science Advances, 2025,11, ads4957. Web Link
Spermidine mediates acetylhypusination of RIPK1 to suppress diabetes onset and progression
T. Zhang, W. Fu, H. Zhang, J. Li, B. Xing, Y. Cai, M. Zhang, X. Liu, C. Qi, L. Qian, X. Hu, H. Zhu, S. Yang, M. Zhang, J. Liu, G. Li, Y. Li, R. Xiang, Z. Qi, J. Hu, Y. Li, C. Zou, Q. Wang, X. Jin, R. Pang, P. Li, J. Liu, Y. Zhang, Z. Wang, Z.-J. Zhu*, B. Shan* and J. Yuan*
Nature Cell Biology, 2024, 26, 2099-2114. Web Link
Met4DX: a unified and versatile data processing tool for multi-dimensional untargeted metabolomics data
Y. Yin, M. Luo, and Z.-J. Zhu*
J. Am. Soc. Mass Spectrom., 2024, 35,2960-2968. Web Link

Fast and broad-coverage lipidomics enabled by ion mobility-mass spectrometry
Y. Cai, X. Chen, F. Ren, H. Wang, Y. Yin, and Z.-J. Zhu*
Analyst, 2024, 149, 5063-5072. Web Link
Plasma metabolomics reveals the shared and distinct metabolic disturbances associated with cardiovascular events in coronary artery disease
J. Lv†, C. Pan†, Y. Cai†, X. Han, C. Wang, J. Ma, J. Pang, F. Xu, S. Wu, T. Kou, F. Ren, Z.-J. Zhu*, T. Zhang*, J. Wang* and Y. Chen*
Nature Communications, 2024,15: 5729. Web Link
Lysophosphatidylcholine binds α-synuclein and prevents its pathological aggregation
C. Zhao†, J. Tu†, C. Wang†, W. Liu, J. Gu, Y. Yin, S. Zhang, D. Li, J. Diao*, Z.-J. Zhu* and C. Liu*
National Science Review, 2024, nwae182. Web Link
Identifying plasma metabolic characteristics of major depressive disorder, bipolar disorder, and schizophrenia in adolescents
B. Yin†, Y. Cai†, T. Teng†, X. Wang, X. Liu, X. Li, Jie Wang, H. Wu, Y. He, F. Ren, T. Kou, Z.-J. Zhu* and X. Zhou*
Translational Psychiatry, 2024, 14: 163. Web Link
Purine salvage–associated metabolites as biomarkers for early diagnosis of esophageal squamous cell carcinoma: a diagnostic model–based study
Y. Sun†, W. Liu†, M. Su, T. Zhang, X. Li, W. Liu, Y. Cai, D. Zhao, M. Yang, Z-J. Zhu*, J. Wang* and J. Yu*
Cell Death Discovery, 2024, 10: 139. Web Link
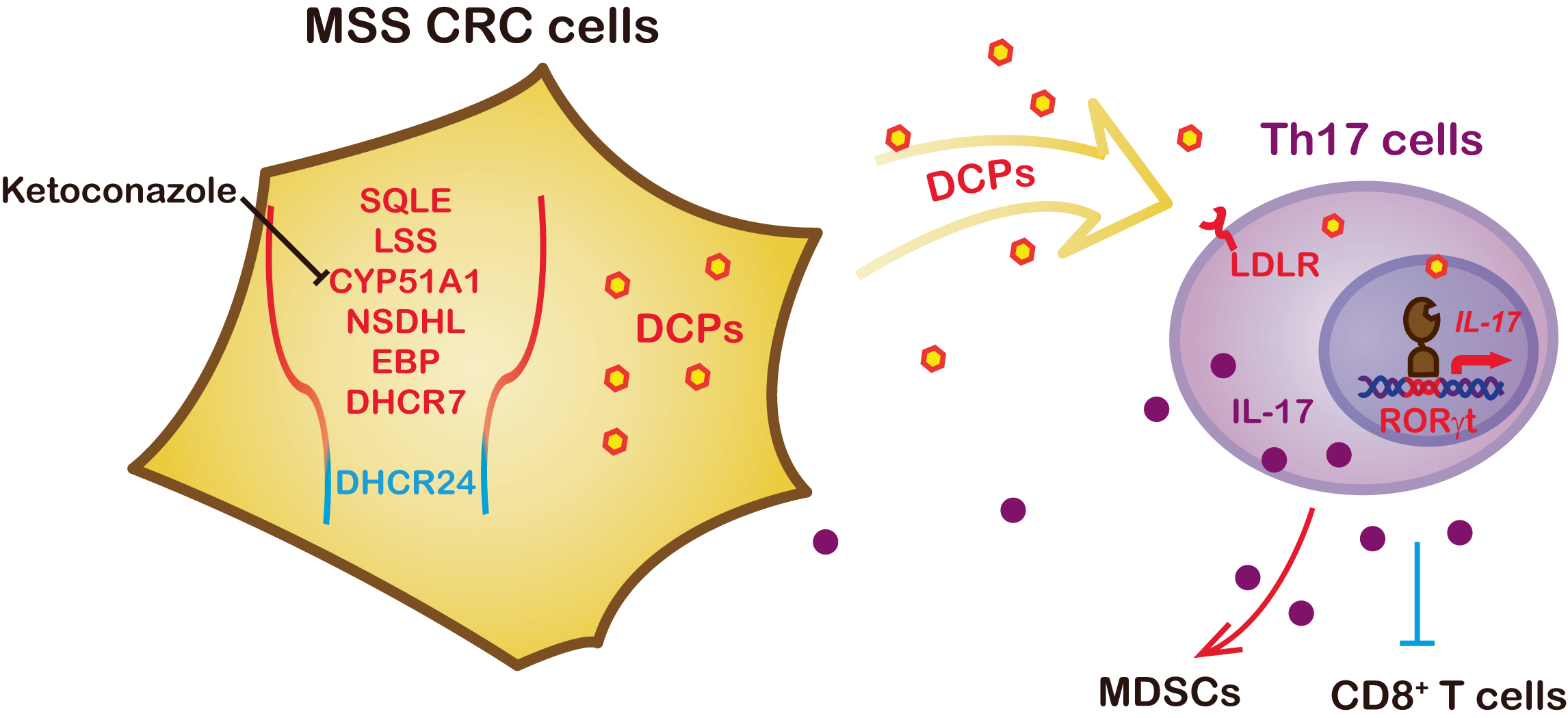
Shaping immune landscape of colorectal cancer by cholesterol metabolites
Y. Bai†, T. Li†, Q. Wang†, W. You†, H. Yang, X. Xu, Z. Li, Y. Zhang, C. Yan, L. Yang, J. Qiu, Y. Liu, S. Chen, D. Wang, B. Huang, K.Liu, B.-L. Song, Z. Wang, K. Li, X. Liu, G. Wang, W. Yang, J. Chen, P Hao, Z. Zhang, Z. Wang*, Z.-J. Zhu* and C. Xu*
EMBO Molecular Medicine, 2024, 16, 334-360. Web Link
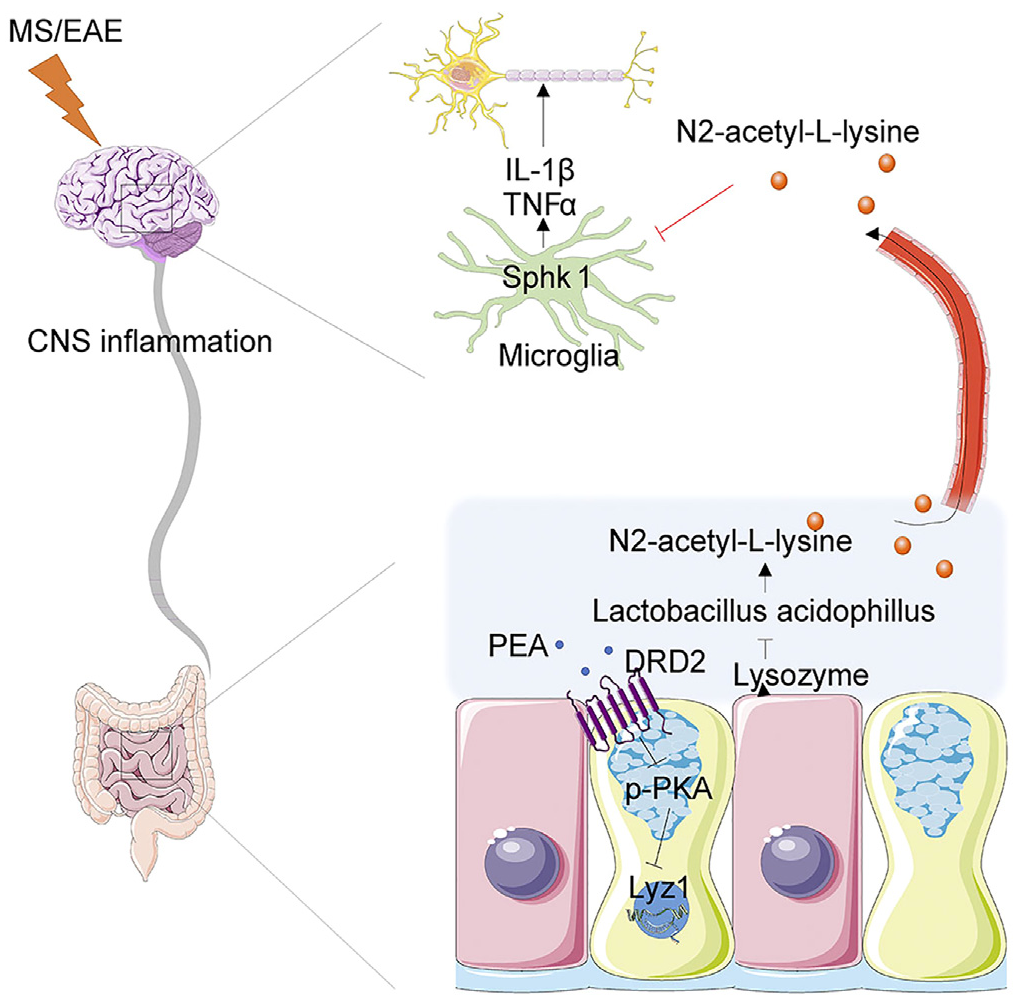
Intestinal epithelial dopamine receptor signaling drives sex-specific disease exacerbation in a mousemodel of multiple sclerosis
H. Peng†, J.-Q. Qiu†, Q. Zhou†, Y. Zhang†, Q. Chen, Y. Yin, W. Su, S. Yu, Y. Wang, Y. Cai, M.Gu, H. Zhang, Q. Sun, G. Hu, Y. Wu, J. Liu, S. Chen*, Z.-J. Zhu*, X. Song*, and J. Zhou*
Immunity, 2023, 56, 2773-2789. Web Link
Allosterically inhibited PFKL via prostaglandin E2 withholds glucose metabolism and ovarian cancer invasiveness
S. Chen, Y. Wu, Y. Gao, C. Wu, Y. Wang, C. Hou, M. Ren, S. Zhang, Q. Zhu, J. Zhang, Y. Yao, M. Huang, Y. B. Qi, X.-S. Liu, T. Horng, H. Wang, D. Ye, Z.-J. Zhu*, S. Zhao* and G. Fan*
Cell Reports, 2023, 42, 113246. Web Link

AllCCS2: Curation of Ion Mobility Collision Cross-Section Atlas for Small Molecules Using Comprehensive Molecular Representations
H. Zhang, M. Luo, H. Wang, F. Ren, Y. Yin, and Z.-J. Zhu*
Analytical Chemistry, 2023, 95, 13913–13921. Web Link
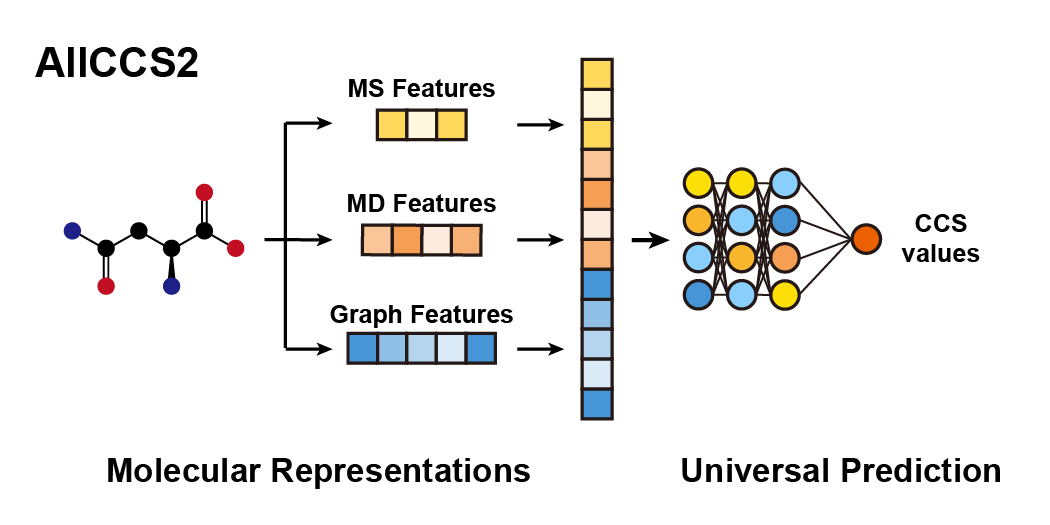
The Oxysterol Receptor EBI2 Links Innate and Adaptive Immunity to Limit IFN Response and Systemic Lupus Erythematosus
F. Zhang, B. Zhang, H. Ding, X. Li, X. Wang, X. Zhang, Q. Liu, Q. Feng, M. Han, L. Chen, L. Qi, D. Yang, X. Li, X. Zhu, Q. Zhao, J. Qiu, Z. J. Zhu, H. Tang, N. Shen*, H. Wang*, and B. Wei*
Advanced Science, 2023: 2207108.
Exhaustion-associated cholesterol deficiency dampens the cytotoxic arm of antitumor immunity
C. Yan, L. Zheng, S. Jiang, H. Yang, J. Guo, L.Jiang, T. Li, H. Zhang, Y. Bai, Y. Lou, Q. Zhang, T. Liang, W.g Schamel, H. Wang, W. Yang, G. Wang, Z.-J. Zhu, B.-L. Song*, and C. Xu*
Cancer Cell, 2023, 41, 1276-1293. Web Link
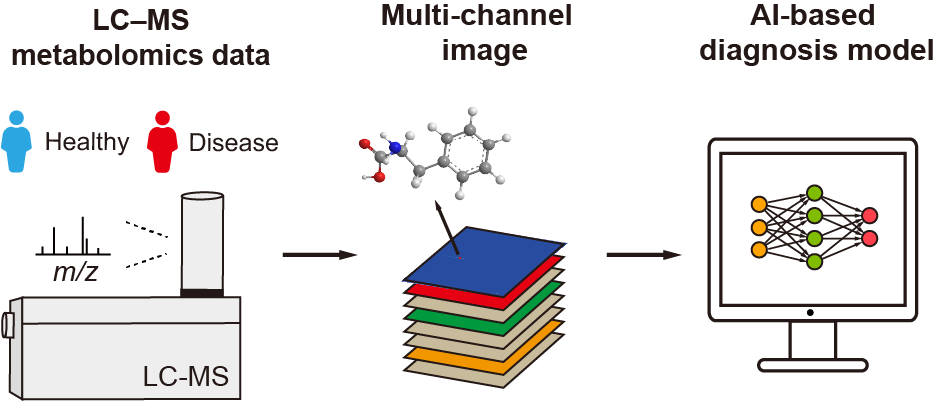
Encoding LC−MS-based Untargeted Metabolomics Data into Images towards AI-based Clinical Diagnosis
H. Wang, Y. Yin,and Z.-J. Zhu*
Analytical Chemistry, 2023, 95, 6533-6541. Web Link
A Mass Spectrum-oriented Computational Method for Ion Mobility-resolved Untargeted Metabolomics
M. Luo, Y. Yin, Z. Zhou, H. Zhang, X. Chen, H. Wang, and Z.-J. Zhu*
Nature Communications, 2023, 14: 1813. Web Link

Selective Covalent Targeting of Pyruvate Kinase M2 Using Arsenous Warheads
J. Wang, S. Zhou, ... W. Liu, J. Zhang, C. Peng, Z.-J. Zhu, M. Huang, Y. Li*, G. Zhuang*, and L. Tan*
Journal of Medicinal Chemistry, 2023, 66, 2608-2621. Web Link
ONE-seq: epitranscriptome and gene-specific profiling of NAD-capped RNA
K. Niu, J. Zhang, S. Ge, D. Li, K. Sun, Y. You, J. Qiu, K. Wang, X. Wang, R. Liu, Y. Liu, B. Li, Z.-J. Zhu, L. Qu*, H.Jiang*, and N. Liu*
Nucleic Acids Research, 2023, 51: e12. Web Link
Advanced Analytical and Informatic Strategies for Metabolite Annotation in Untargeted Metabolomics
Y. Cai, Z. Zhou, and Z.-J. Zhu*
Trends in Analytical Chemistry, 2023, 158: 116903. (Invited Review) Web Link
Serum Metabolic Traits Reveal Therapeutic Toxicities and Responses of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer
H. Wang†, H. Jia†, Y. Gao, H. Zhang, J. Fan, L. Zhang, F. Ren, Y. Yin, Y. Cai*, J. Zhu*, and Z.-J. Zhu*
Nature Communications, 2022, 13: 7802. Web Link
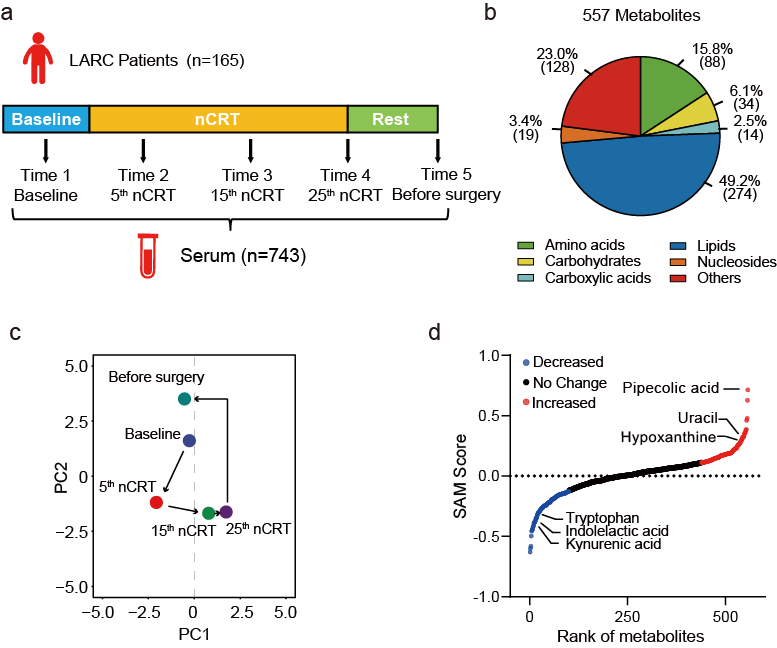
Metabolite Annotation from Knowns to Unknowns through Knowledge-guided Multi-layer Metabolic Networking
Z. Zhou†, M. Luo†, H. Zhang, Y. Yin, Y. Cai, and Z.-J. Zhu* (Corresponding author)
Nature Communications, 2022, 13: 6656. Web Link
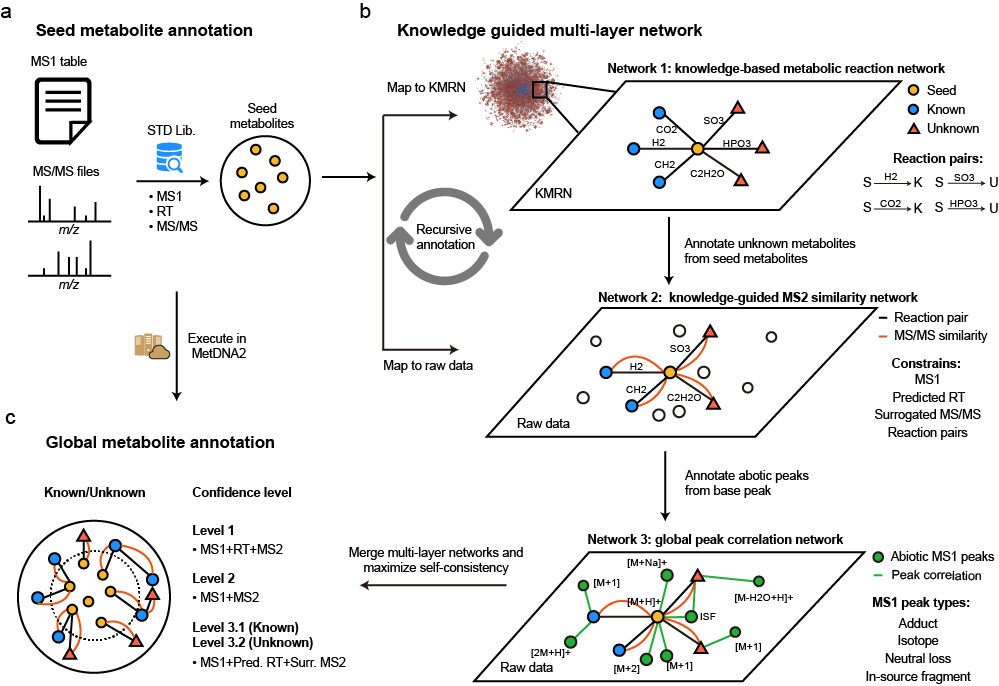
Four-dimensional Untargeted Profiling of N-acylethanolamine Lipids in the Mouse Brain Using Ion Mobility-Mass Spectrometry
W. Liu, W. Zhang*, T. Li, Z. Zhou, M. Luo, X. Chen, Y. Cai*, and Z.-J. Zhu* (Corresponding author),
Analytical Chemistry, 2022, 94, 12472-12480. Web Link
Quantitative Acetylomics Reveals Dynamics of Protein Lysine Acetylation in Mouse Livers During Aging and Upon the Treatment of Nicotinamide Mononucleotide
J. Li, Y. Cao, K. Niu, J. Qiu, H. Wang, Y. You, D. Li, Y. Luo, Z. J. Zhu, Y. Zhang*, and N. Liu*
Molecular and Cellular Proteomics, 2022, 21, 100276.
Global Stable-isotope Tracing Metabolomics Reveals System-wide Metabolic Alternations in Aging Drosophila
R. Wang, Y. Yin, J. Li, H. Wang, W. Lv, Y. Gao, T. Wang, Y. Zhong, Z. Zhou, Y. Cai, X. Su, N. Liu*, and Z.-J. Zhu* (Corresponding author)
Nature Communications, 2022, 13: 3518. Web Link
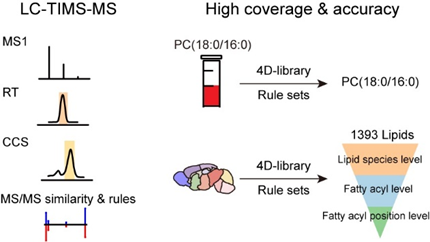
Trapped Ion Mobility Spectrometry-Mass Spectrometry Improves the Coverage and Accuracy of Four-dimensional Untargeted Lipidomics
X. Chen†, Y. Yin†, M. Luo, Z. Zhou, Y. Cai*, and Z.-J. Zhu* (Corresponding author)
Analytica Chimica Acta, 2022, 1210: 339886. Web Link
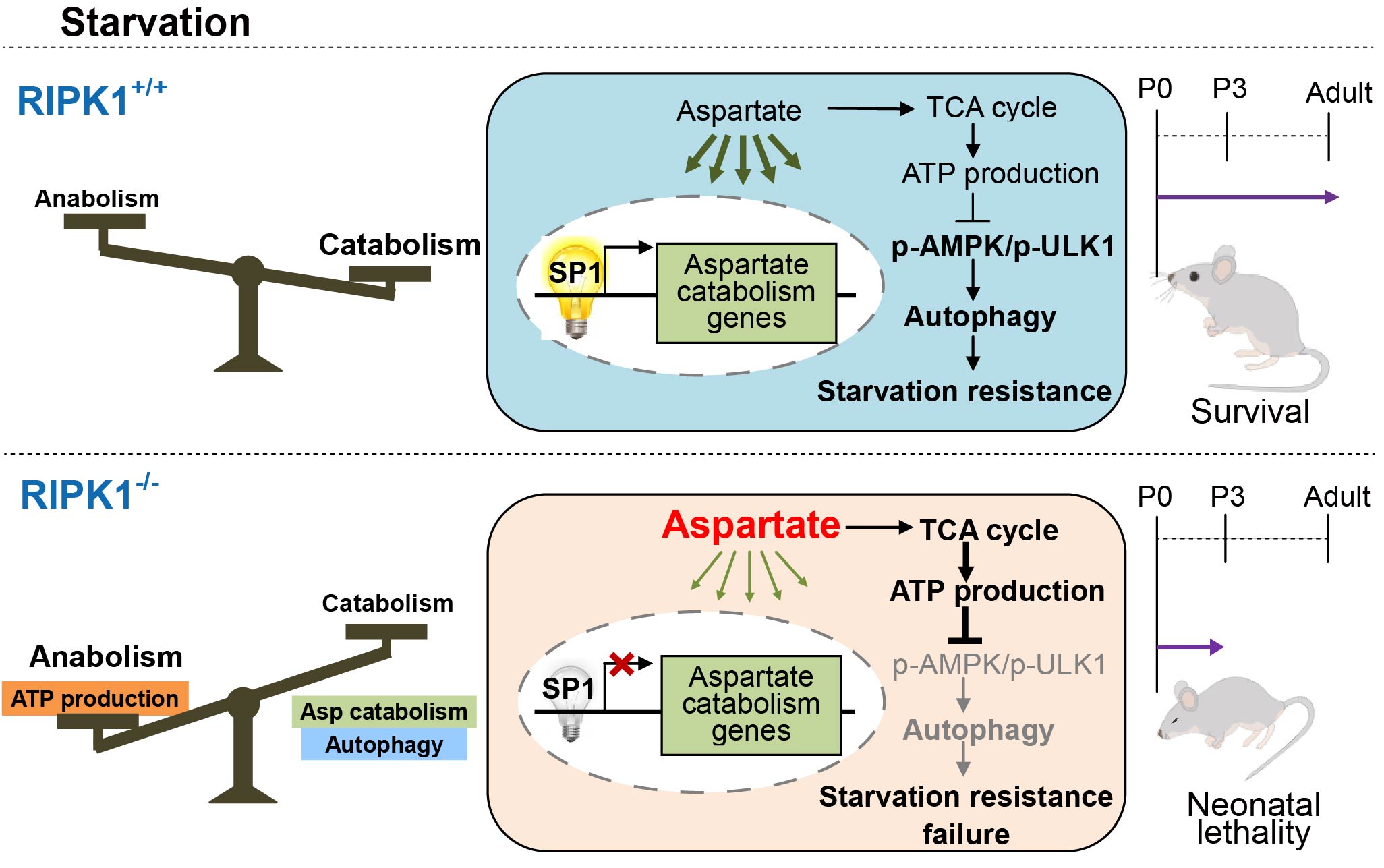
RIPK1 Regulates Starvation Resistance by Modulating Aspartate Catabolism
X. Mei†, Y. Guo†, Z. Xie†, Y. Zhong, X. Wu, D. Xu, Y. Li, N. Liu, and Z.-J. Zhu* (corresponding author)
Nature Communications, 2021, 12: 6144. Web link
WaveICA 2.0: a novel batch effect removal method for untargeted metabolomics data without using batch information
K. Deng, F. Zhao, Z. Rong, L. Cao, L. Zhang, K.Li*, Y. Hou*, and Z.-J. Zhu*
Metabolomics, 2021, 17: 87
metID: a R package for automatable compound annotation for LC−MS-based data
X. Shen, S. Wu, L. Liang, S. Chen, K. Contrepois, Z.-J. Zhu*, and M. Snyder*
Bioinformatics, 2021, btab583. Web link
NEK1-mediated Retromer Trafficking Promotes Blood–Brain Barrier Integrity by Regulating Glucose Metabolism and RIPK1 Activation
H. Wang, W. Qi, C. Zou, Z. Xie, M. Zhang, M. Naito, L. Mifflin, Z. Liu, A. Najafov, H. Pan, B.Shan, Y. Li, Z.-J. Zhu, and J. Yuan*
Nature Communications, 2021, 12: 4826. Web link
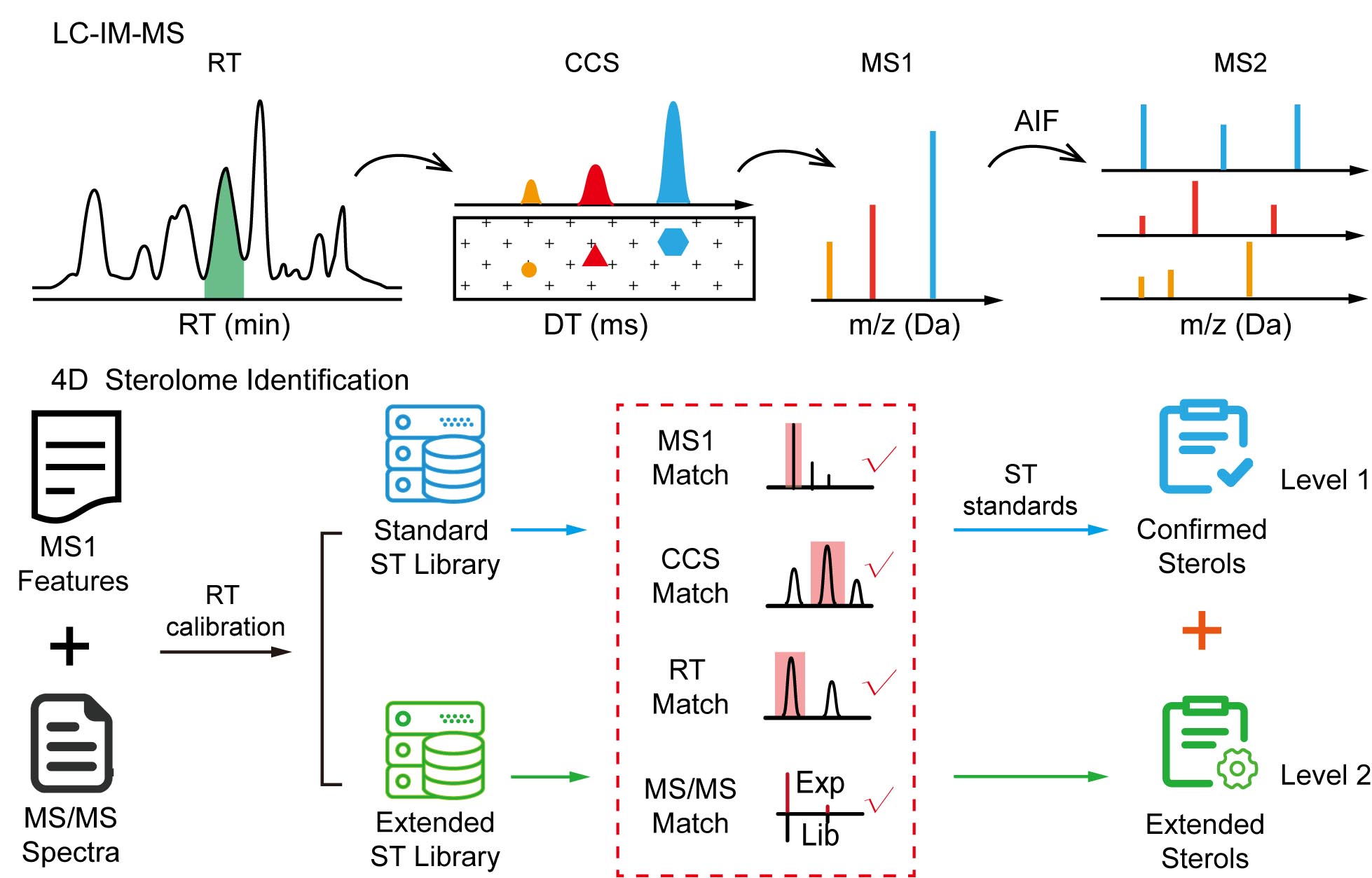
Ion Mobility-based Sterolomics Reveals Spatially and Temporally Distinctive Sterol Lipids in the Mouse Brain
T. Li, Y. Yin, Z. Zhou, J. Qiu, W. Liu, X. Zhang, K. He, Y. Cai, and Z.-J. Zhu* (corresponding author)
Nature Communications, 2021, 12: 4343. Web Link BrainSterolAtlas
Serum metabolomics identifies dysregulated pathways and potential metabolic biomarkers for hyperuricemia and gout
X. Shen, C. Wang, N. Liang, Z. Liu, X. Li, Z.-J. Zhu, T. Merriman, N. Dalbeth, R.Terkeltaub, C. Li*, and H. Yin*
Arthritis & Rheumatology, 2021, 73, 1738-1748. Web link
A Serum Metabolomics Analysis Reveals A Panel of Screening Metabolic Biomarkers for Esophageal Squamous Cell Carcinoma
J. Lv, J. Wang, X. Shen, J. Liu, D. Zhao, M. Wei, X. Li, B. Fan, Y. Sun, F. Xue, Z.-J. Zhu* (corresponding author), and T. Zhang*
Clinical and Translational Medicine, 2021, 11: e419. Web link
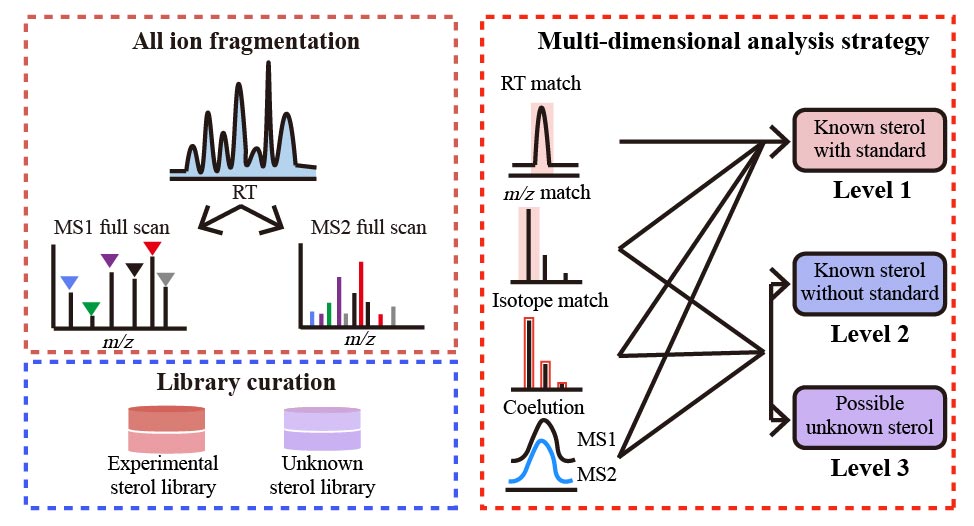
Multi-dimensional Characterization and Identification of Sterols in Untargeted LC-MS Analysis Using All Ion Fragmentation Technology
J. Qiu†, T. Li†, and Z.-J. Zhu* (corresponding author)
Analytica Chimica Acta, 2021, 1142, 108-117. Web Link
Exploring the Protective Effects of Danqi Tongmai Tablet on Acute Myocardial Ischemia Rats by Comprehensive Metabolomics Profiling
Z. Li, J. Hou, Y. Deng, H. Zhi, W. Wu, B. Yan, T. Chen, J. Tu, Z.J. Zhu, W. Wu*, and D. Guo*,
Phytomedicine, 2020, 74, 152918. Web Link
Subacute Toxicity Study of Nicotinamide Mononucleotide via Oral Administration
Y. You, Y. Gao, H. Wang, J. Li, X. Zhang, Z.-J. Zhu, and N. Liu*
Frontiers in Pharmacology, 2020, 11, Article no. 604404. Web Link
Development of A Combined Strategy for Accurate Lipid Structural Identification and Quantification in Ion-Mobility Mass Spectrometry based Untargeted Lipidomics
X. Chen†, Y. Yin†, Z. Zhou, T. Li, and Z.-J. Zhu* (corresponding author)
Analytica Chimica Acta, 2020, 1136, 115-124. Web Link Invited contribution for the Special Issue "Analytical Lipidomics"
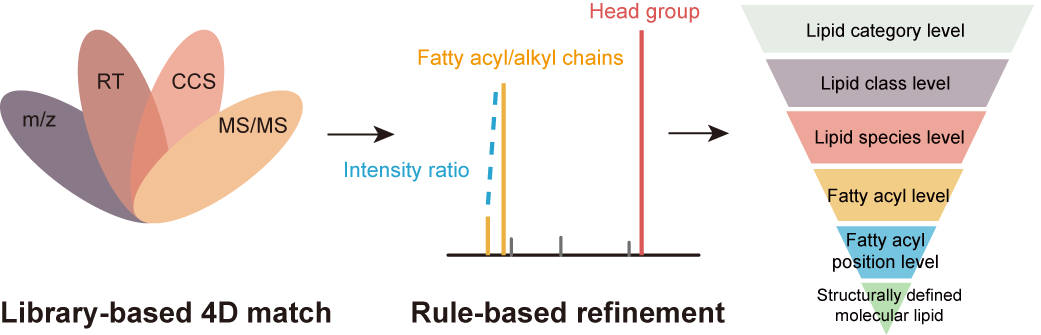
Ion Mobility Collision Cross-Section Atlas for Known and Unknown Metabolite Annotation in Untargeted Metabolomics
Z. Zhou, M. Luo, X. Chen, Y. Yin, X. Xiong, R. Wang, and Z.-J. Zhu* (corresponding author)
Nature Communications, 2020, 11: 4334. Web Link AllCCS Web

A lipidome atlas in MS-DIAL 4
H. Tsugawa , K. Ikeda, M. Takahashi , A. Satoh, Y. Mori, H. Uchino, N. Okahashi, Y. Yamada, I. Tada, P. Bonini, Y. Higashi, Y. Okazaki, Z. Zhou, Z.-J. Zhu, J. Koelmel, T. Cajka , O. Fiehn, K. Saito, M. Arita, and M. Arita*
Nature Biotechnology, 2020, 38, 1159-1163. Web Link
The Application of Ion Mobility-Mass Spectrometry in Untargeted Metabolomics: from Separation to Identification
M. Luo, Z. Zhou, and Z.-J. Zhu* (corresponding author)
Journal of Analysis and Testing, 2020, 4, 163-174. Web Link Invited review for the Special Issue "Metabolomics: state of art in method development and applications"
NormAE: Deep Adversarial Learning Model to Remove Batch Effects in Liquid Chromatography Mass Spectrometry-Based Metabolomics Data
Z. Rong, Q. Tan, L. Cao, L. Zhang, K. Deng, Y. Huang, Z.-J. Zhu, Z. Li, and K. Li*
Analytical Chemistry, 2020, 92, 5082-5090. Web Link
Different Regions of Synaptic Vesicle Membrane Regulate VAMP2 Conformation for the SNARE Assembly
C. Wang, J. Tu, S. Zhang, B. Cai, Z. Liu, S. Hou, Q. Zhong, X. Hu, W. Liu, G. Li, Z. Liu, L. He, J. Diao, Z.-J. Zhu, Dan Li *, and C. Liu*
Nature Communications, 2020, 11: 1531. Web link
Overview of Tandem Mass Spectral and Metabolite Databases for Metabolite Identification in Metabolomics
Z. Yi, and Z.-J. Zhu* (corresponding author)
Methods in Molecular Biology, 2020, 2124, 139-148. Web Link An invited book chapter for Computational Methods and Data Analysis for Metabolomics
Daily Oscillation of the Excitation-Inhibition Balance in Visual Cortical Circuits
M.C.D. Bridi, F. J. Zong, X. Min, N. Luo, T. Tran, J. Qiu, D. Severin, X.T. Zhang, G. Wang, Z.-J. Zhu, K.W. He,* and A. Kirkwood*
Neuron, 2020, 105, 621-629.
The Use of LipidIMMS Analyzer for Lipid Identification in Ion Mobility-Mass Spectrometry-Based Untargeted Lipidomics
X. Chen, Z. Zhou, and Z.-J. Zhu* (Corresponding author)
Methods in Molecular Biology, 2020, 2084, 269-282. Web Link An invited book chapter for Ion Mobility-Mass Spectrometry: Methods and Protocols
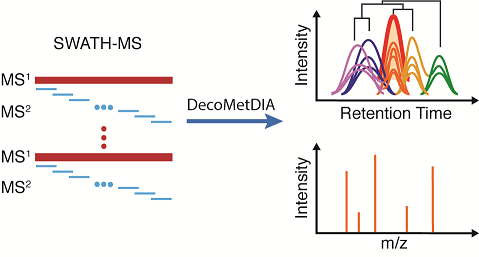
DecoMetDIA: Deconvolution of Multiplexed MS/MS Spectra for Metabolite Identification in SWATH-MS based Untargeted Metabolomics
Y. Yin†, R. Wang †, Y. Cai, Z. Wang, and Z.-J. Zhu* (Corresponding author)
Analytical Chemistry, 2019, 91, 11897-11904. Web Link
A Vitamin-C-derived DNA Modification Catalysed by An Algal TET Homologue
J. Xue†, G. Chen†, F. Hao†, H. Chen†, Z. Fang, F.-F. Chen, B. Pang, Q. Yang, X. Wei, Q. Fan, C. Xin, J. Zhao, X. Deng, B. Wang, X. Zhang, Y. Chu, H. Tang, H. Yin, W. Ma, L. Chen, J. Ding, E. Weinhold, R. M. Kohli, W. Liu, Z.-J. Zhu, K. Huang*, H. Tang* , and G.-L. Xu*
Nature, 2019, 569, 581–585. Web Link
Metabolic Reaction Network-based Recursive Metabolite Annotation for Untargeted Metabolomics
X. Shen, R. Wang, X. Xiong, Y. Yin, Y. Cai, Z. Ma, N. Liu, and Z.-J. Zhu* (Corresponding author)
Nature Communications, 2019, 10: 1516. Web Link MetDNA Webserver

The Emerging Role of Ion Mobility-Mass Spectrometry in Lipidomics to Facilitate Lipid Separation and Identification
J. Tu†, Z. Zhou†,T. Li†, and Z.-J. Zhu* (Corresponding Author)
Trends in Analytical Chemistry, 2019, 116, 332-339. Web Link An invited contribution to the special issue of “Ion Mobility Spectrometry: From Fundamentals to Applications”.
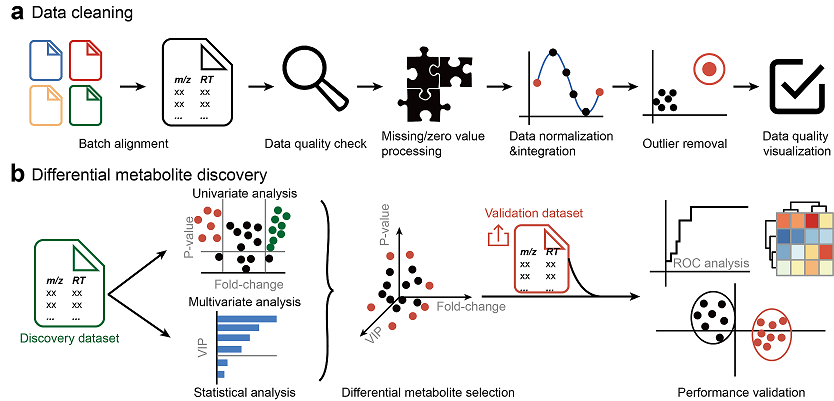
MetFlow: An Interactive and Integrated Workflow for Metabolomics Data Cleaning and Differential Metabolite Discovery
X. Shen, and Z.-J. Zhu*(Corresponding author)
Bioinformatics, 2019, 35, 2870-2872. Web Link Web Server Link
Advancing Untargeted Metabolomics Using Data Independent Acquisition Mass Spectrometry Technology
R. Wang†, Y. Yin†, and Z.-J. Zhu* (Corresponding author)
Analytical and Bioanalytical Chemistry, 2019, 411, 4235-4250. Web Link An invited contribution to the special issue of “Young Investigators in Analytical and Bioanalytical Science”.
WaveICA: A novel algorithm to remove batch effects for large-scale untargeted metabolomics data based on wavelet analysis
K. Deng, F. Zhang, Q. Tan, Y. Huang, W. Song, Z. Rong, Z.-J. Zhu, K. Li*, and Z. Li*,
Analytica Chimica Acta, 2019, 1061, 60-69.
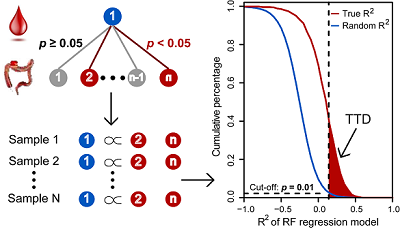
Development of A Correlative Strategy to Discover Colorectal Tumor Tissue Derived Metabolite Biomarkers in Plasma Using Untargeted Metabolomics
Z. Wang†, B. Cui†, F. Zhang, Y. Yang, X. Shen, Z. Li, W. Zhao, Y. Zhang, K. Deng, Z. Rong, K. Yang, X. Yu, K. Li*, P. Han*, and Z.-J. Zhu*
Analytical Chemistry, 2019, 91, 2401-2408. Web Link (†, Co-first authors; *, Co-Corresponding authors)
LipidIMMS Analyzer: Integrating Multi-dimensional Information to Support Lipid Identification in Ion Mobility–Mass Spectrometry based Lipidomics
Z. Zhou, X. Shen, X. Chen, J. Tu, X. Xiong, and Z.-J. Zhu* (Corresponding author)
Bioinformatics, 2019, 35,698-700. Web Link LipidIMMS Web Server Link
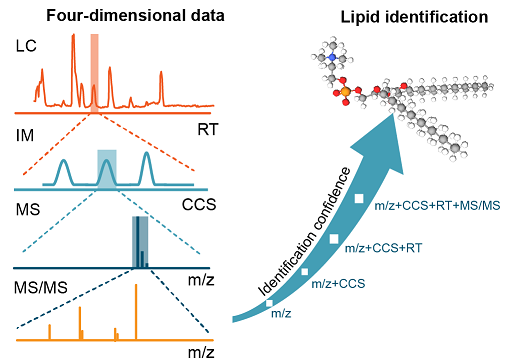
A High-Throughput Targeted Metabolomics Workflow for the Detection of 200 Polar Metabolites in Central Carbon Metabolism
Y. Cai, and Z.-J. Zhu* (Corresponding author)
Methods in Molecular Biology, 2019, 1859,263-274. Web Link An invited book chapter for Microbial Metabolomics
Metabolomics Approach for Predicting Response to Neoadjuvant Chemotherapy for Colorectal Cancer
K. Yang, F. Zhang, P. Han, Z. Z. Wang, K. Deng, Y. Y. Zhang, W. W. Zhao, W. Song, Y. Q. Cai, K. Li*, B. B. Cui*, and Z.-J. Zhu* (Corresponding author)
Metabolomics, 2018,14: 110.
Stable-isotope Labeled Metabolic Analysis in Drosophila melanogaster: From Experimental Setup to Data Analysis
Y. Cai, N. Liu*, and Z.-J. Zhu* (Corresponding author)
Bio-protocol, 2018, 8: e3015. Web Link
Predicting the Pathological Response to Neoadjuvant Chemoradiation Using Untargeted Metabolomics in Locally Advanced Rectal Cancer
H. Jia†, X. Shen†, Y. Guan, M. Xu, J. Tu, M. Mo, L. Xie, J. Yuan, J. Zhu*, and Z.-J. Zhu* (Corresponding author)
Radiotherapy and Oncology, 2018, 128, 548-556.
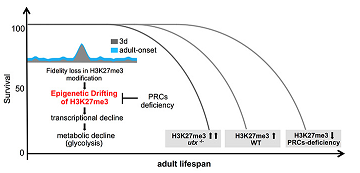
Epigenetic Drift of H3K27me3 in Aging Links Glycolysis to Healthy Longevity in Drosophila
Z. Ma†, H. Wang†, Y. Cai†, H. Wang†, K. Niu, X. Wu, H. Ma, Y. Yang, W. Tong, F. Liu, Z. Liu, Y. Zhang, R. Liu, Z.-J. Zhu*, and N. Liu*
eLife, 2018, 7: e35368 Web Link (†, Co-first authors; *, Co-Corresponding authors)Preprint at BioRxiv (doi.org/10.1101/247726)
--- This paper is highlighted by Science: "Epigenetics, aging, and glycolysis" Web Link
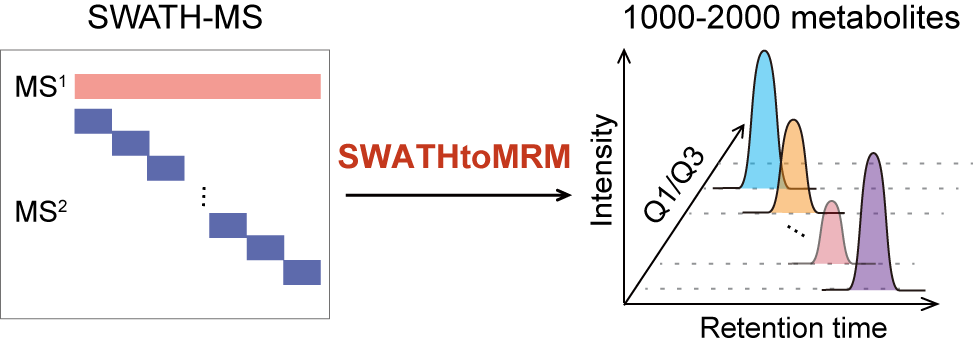
SWATHtoMRM: Development of High-Coverage Targeted Metabolomics Method Using SWATH Technology for Biomarker Discovery
H. Zha†, Y. Cai†, Y. Yin†, Z. Wang, K. Li, and Z.-J. Zhu* (Corresponding author)
Analytical Chemistry, 2018, 90, 4062-4070. Web Link
Absolute Quantitative Lipidomics Reveals Lipidome-Wide Alterations in Aging Brain
J. Tu, Y. Yin, M. Xu, R. Wang, and Z.-J. Zhu*(Corresponding author)
Metabolomics, 2018,14: 5. Web Link
Advancing the Large-Scale CCS Database for Metabolomics and Lipidomics at the Machine-Learning Era
Z. Zhou†, J. Tu†, and Z.-J. Zhu* (Corresponding author)
Current Opinion in Chemical Biology, 2018, 42, 34-41. Web Link Recommended by F1000
CLOCK Acetylates ASS1 to Drive Circadian Rhythm of Ureagenesis,
R. Lin*, Y. Mo, H. H. Zha, Z. Qu, P. Xie, Z.-J. Zhu, Y. Xu, Y. Xiong*, K.-L.Guan*
Molecular Cell, 2017, 68, 198–209.
Proteome-Wide Analysis of N-Glycosylation Stoichiometry Using SWATH Technology
X. Yang, Z. Wang, L. Guo, Z.-J. Zhu, and Y. Zhang*
Journal of Proteome Research, 2017, 16, 3830–3840.
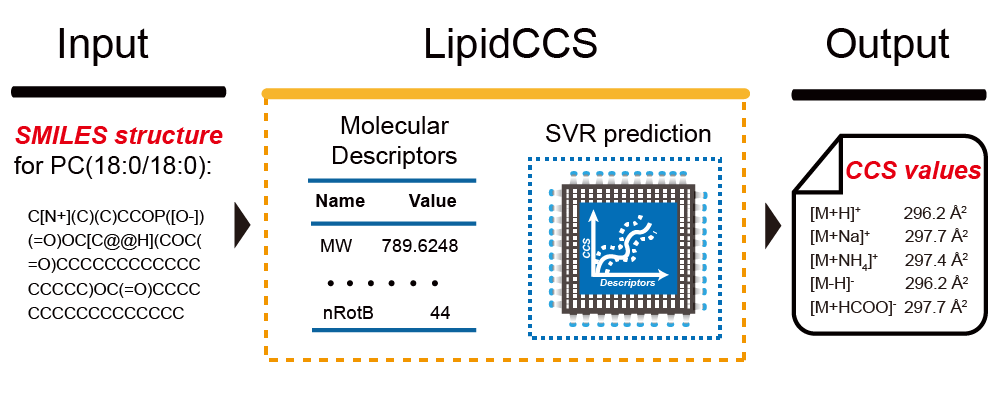
LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision to Support Ion Mobility-Mass Spectrometry based Lipidomics
Z. Zhou, J. Tu, X. Xiong, X. Shen, and Z.-J. Zhu* (Corresponding author)
Analytical Chemistry, 2017, 89, 9559–9566. Web Link Web Link for LipidCCS Server
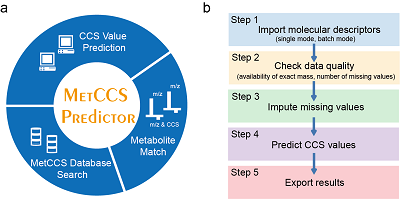
MetCCS Predictor: A Web Server for Predicting Collision Cross-Section Values of Metabolites in Metabolomics
Z. Zhou, X. Xiong, and Z.-J. Zhu* (Corresponding author)
Bioinformatics, 2017, 33, 2235-2237. Web Link for Publication Web Link for MetCCS Server
Comprehensive Metabolomics Identified Lipid Peroxidation as A Prominent Feature in Human Plasma of Patients with Coronary Heart Diseases
J. Lu, B. Chen, T. T. Chen, S. Guo, X. Xue, Q. Chen, M. Zhao, L. Xia, Z.-J. Zhu, L. Zheng*, H. Yin*
Redoxy Biology, 2017, 12, 899-907. Web Link
Discovery of novel 1,2,3,4-tetrahydrobenzo[4, 5]thieno[2, 3-c]pyridine derivatives as potent and selective CYP17 inhibitors
M. Wang, Y. Fang, S. Gu, F. F. Chen, Z.-J. Zhu, X. Sun*, J. Zhu*
European Journal of Medicinal Chemistry, 2017, 132, 157-172.
Normalization and Integration of Large-Scale Metabolomics Data Using Support Vector Regression
X. Shen, X. Gong, Y. Cai, Y. Guo, J. Tu, H. Li, T.Zhang, J. Wang, F. Xue, and Z.-J. Zhu* (Corresponding author)
Metabolomics, 2016, 12: 89 Web Link
Serum Metabolomics for Early Diagnosis of Esophageal Squamous Cell Carcinoma by UHPLC-QTOF/MS,
J. Wang*,†, T. Zhang†, X. Shen†, J. Liu, D. Zhao, Y. Sun, L. Wang, Y. Liu, X. Gong, Y. Liu, Z.-J. Zhu*, and F. Xue,*
Metabolomics, 2016, 12: 116 Web Link
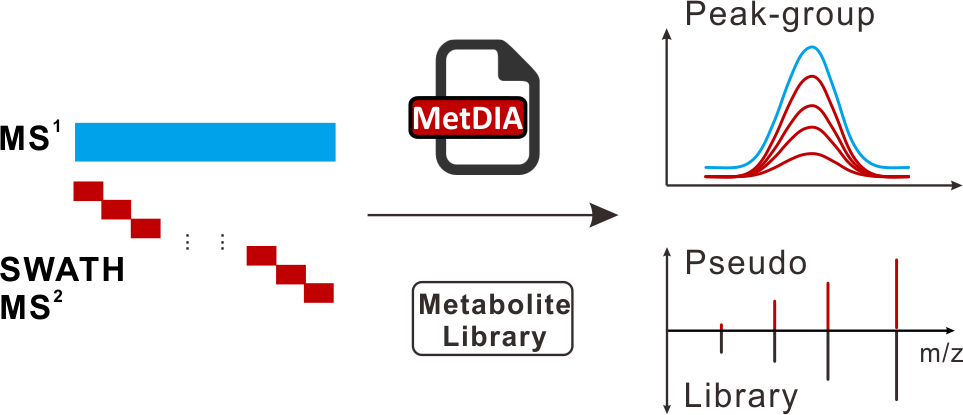
MetDIA: Targeted Metabolite Extraction of Multiplexed MS/MS Spectra Generated by Data-Independent Acquisition
H. Li, Y. Cai, Y. Guo, F. Chen, and Z.-J. Zhu* (Corresponding author)
Analytical Chemistry, 2016, 88, 8757-8764. Web Link
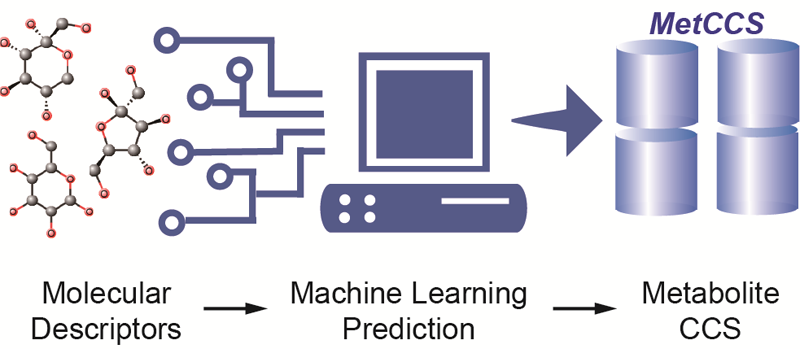
Large-Scale Prediction of Collision Cross-Section Values for Metabolites in Ion Mobility - Mass Spectrometry
Z. Zhou, X. Shen, J. Tu, and Z.-J. Zhu* (Corresponding author)
Analytical Chemistry, 2016, 88, 11084-11091. Web Link
Y. Cai, K. Weng, Y. Guo, J. Peng, and Z.-J. Zhu* (Corresponding author), An Integrated Targeted Metabolomic Platform for High-Throughput Metabolite Profiling and Automated Data Processing, Metabolomics, 2015,11,1575-1586. Web Link
H.- G. Xia, A. Najafov,. J.Geng,. L. Galan-Acosta, X. Han,. Y. Guo, B. Shan, Y. Zhang, E. Norberg, T. Zhang, L.Pan, J.Liu, J. L. Coloff, D. Ofengeim, H. Zhu, K. Wu, Y. Cai, J. R. Yates, Z.-J. Zhu, J. Yuan, and H. Vakifahmetoglu-Norberg, Degradation of HK2 by Chaperone-Mediated Autophagy Promotes Metabolic Catastrophe and Cell Death, Journal of Cell Biology, 2015, 210,705-716.
M. E. Kurczy, Z.-J. Zhu, ... 20 other authors..., G. Siuzdak*, Comprehensive Bioimaging with Fluorinated Nanoparticles Using Breathable Liquids, Nature Communications, 2015, 6, 5998.
J. Ivanisevic, D. Elias, H. Deguchi, P.M. Averell, M. Kurczy, C. H. Johnson, R. Tautenhahn, Z.-J. Zhu, J. Watrous, M.Jain, J.Griffin, G.J. Patti and G. Siuzdak*, Arteriovenous Blood Metabolomics: A Readout of Intra-Tissue Metabostasis, Scientific Reports, 2015, 5, 12757.
C. Kim, G. Y. Tonga,B. Yan, C. S. Kim,S. T. Kim, M. H. Park, Z.-J. Zhu, B. Duncan,B. Creran, and V. M. Rotello*, Regulating Exocytosis of Nanoparticles via Host-guest Chemistry,Organic and Biomolecular Chemistry, 2015,13, 2474-2479.
J. Ivanisevic, Z.-J. Zhu (Co-first author), L. Plate, R. Tautenhahn, S. Chen, P.J. O’Brien, C.H. Johnson, M. A. Marletta, G.J. Patti, and G. Siuzdak*, Toward 'Omic' Scale Metabolite Profiling: A Dual Separation – Mass Spectrometry Approach for Coverage of Lipid and Central Carbon Metabolism, Analytical Chemistry, 2013, 85, 6876–6884.
Z.-J. Zhu, A.W. Schultz, J. Wang, C. H. Johnson, G. J. Patti, and G. Siuzdak*, Liquid Chromatography Quadrupole Time-of-Flight Characterization of Metabolites Guided by the METLIN Database, Nature Protocols, 2013, 8, 451-460.
R. Tang, C. S. Kim, D. J. Solfiell, S. Rana, R. Mout, E. M. Velazquez-Delgado, A. Chompoosor, Y. Jeong, B. Yan, Z.-J. Zhu, C. Kim, J. A. Hardy, and V. M. Rotello*, Direct Delivery of Functional Proteins and Enzymes to the Cytosol Using Nanoparticle-Stabilized Nanocapsules, ACS Nano, 2013, 7, 6667-6673.
B. Yan, Y. Jeong, L. A. Mercante, G. Y. Tonga, C. Kim, Z.-J. Zhu, R. W. Vachet*, and V. M. Rotello*, Characterization of surface ligands on functionalized magnetic nanoparticles using laser desorption/ionization mass spectrometry (LDI-MS), Nanoscale, 2013, 5, 5063-5066.
R. Tautenhahn, K. Cho, W. Uritboonthai, Z.-J. Zhu, G. J. Patti, and G. Siuzdak*, An Accelerated Workflow for Untargeted Metabolomics Using the METLIN Database, Nature Biotechnology, 2012, 30, 826-828.
Z.-J. Zhu, H. Wang, B. Yan, H. Zheng, Y. Jiang, O. R. Miranda, V. M. Rotello*, B. Xing*, and R. W. Vachet*, Effect of Surface Charge on the Uptake and Distribution of Gold Nanoparticles in Four Plant Species, Environmental Science and Technology, 2012, 22,12391-12398.
Z.-J. Zhu, T. Posati, D. F. Moyano, R. Tang, B. Yan, R. W. Vachet*, and V. M. Rotello*, The Interplay of Monolayer Structure and Serum Protein Interactions on the Cellular Uptake of Gold Nanoparticles, Small, 2012, 8, 2659-2663.
Z.-J. Zhu, R. Tang, Y.-C. Yeh, O. R. Miranda, V. M. Rotello*, and R. W. Vachet*, Determination of the Intracellular Stability of Gold Nanoparticle Monolayers Using Mass Spectrometry, Anal. Chem., 2012, 84, 4321-4326.
Z.-J. Zhu, Y.-C. Yeh, R. Tang, R., B. Yan, Tamayo, J., R. W. Vachet*, and V. M. Rotello*, Stability of Quantum Dots in Live Cells, Nature Chemistry, 2011, 3, 963-968.
O. R. Miranda, X. Li, L. Garcia-Gonzalez, Z.-J. Zhu, B. Yan, U. H. F. Bunz, and V. M. Rotello, Colorimetric Bacteria Sensing Using a Supramolecular Enzyme-Nanoparticle Biosensor, J. Am. Chem. Soc., 2011, 133, 9650-9653.
X.-C. Yang, B. Samanta, S. S. Agasti, Y. Jeong, Z.-J. Zhu, S. Rana, O. R. Miranda, and V. M. Rotello*, Drug Delivery Using Nanoparticle‐Stabilized Nanocapsules, Angew. Chem., Int. Ed. 2011, 50, 477-481.
C. K. Kim, S. S. Agasti, Z.-J. Zhu, L. Isaacs, and V. M. Rotello*, Host-Guest Chemistry inside Living Cells: Recognition-Mediated Activation of Therapeutic Gold Nanoparticles, Nature Chemistry, 2010, 2, 962-966.
Z.-J. Zhu, R. Carboni, M. J. Quercio Jr., B. Yan, O. R. Miranda, D. L. Anderton, K. F. Arcaro*, V. M. Rotello*, and R. W. Vachet*, Surface Properties Dictate Uptake, Distribution, Excretion, and Toxicity of Nanoparticles in Fish, Small, 2010, 6, 2261-2265.
A. Chompoosor, K. Saha, P. S. Ghosh, O. R. Miranda, Z.-J. Zhu, K. F. Arcaro, and V. M. Rotello*, The Role of Surface Functionality on Acute Cytotoxicity, ROS Generation and DNA Damage by Cationic Gold Nanoparticles, Small, 2010, 6, 2246-2249.
P. Ghosh, X. Yang, R. Arvizo, Z.-J. Zhu, H. Mo, and V. M. Rotello*, Intracellular Delivery of a Membrane-Impermeable Enzyme in Active Form Using Functionalized Gold Nanoparticles, J. Am. Chem. Soc., 2010, 132, 2642–2645.
B. Yan, Z.-J. Zhu, O. R. Miranda, A. Chompoosor, V. M. Rotello*, and R. W. Vachet*, Laser Desorption/Ionization Mass Spectrometry Analysis of Monolayer-protected Gold Nanoparticles, Anal. Bioanal. Chem., 2010, 396, 1025-1035.
Z.-J. Zhu, V. M. Rotello*, and R. W. Vachet*, Engineered Nanoparticle Surfaces for Improved Mass Spectrometric Analyses, Analyst, 2009, 134, 2183-2188.
C. K. Kim, P. Ghosh, C. Pagliuca, Z.-J. Zhu, S. Menichetti, and V. M. Rotello*, Entrapment of Hydrophobic Drugs in Nanoparticle Monolayers with Efficient Release into Cancer Cells, J. Am. Chem. Soc., 2009, 131, 1360–1361.
Z.-J. Zhu, P. S. Ghosh, O. R. Miranda, R. W. Vachet*, and V. M. Rotello*, Multiplexed Screening of Cellular Uptake of Gold Nanoparticles Using Laser Desorption/Ionization Mass Spectrometry, J. Am. Chem. Soc., 2008, 130, 14139-14143.
W.-Z. Jia, K. Wang, Z.-J. Zhu, H.-T. Song, and X.-H. Xia*, One-Step Immobilization of Glucose Oxidase in a Silica Matrix on a Pt Electrode by an Electrochemically Induced Sol-Gel Process, Langmuir, 2007, 23, 11896-11900.