
MetCCS: Large-scale Prediction of Collision Cross-Section (CCS) Values for Metabolites in Ion Mobility — Mass Spectrometry
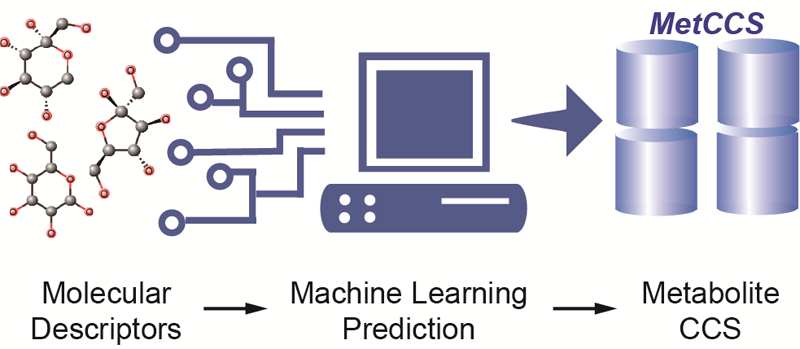
Brief introduction:The rapid development of metabolomics has significantly advanced health and disease related research. However, metabolite identification remains a major analytical challenge for untargeted metabolomics. While the use of collision cross-section (CCS) values obtained in ion mobility - mass spectrometry (IM-MS) effectively increases identification confidence of metabolites, it is restricted by the limited number of available CCS values for metabolites. Here, we demonstrated the use of a machine-learning algorithm called support vector regression (SVR) to develop a prediction method that utilized 14 common molecular descriptors to predict CCS values for metabolites. In this work, we first experimentally measured CCS values (ΩN2) of ~400 metabolites in nitrogen buffer gas, and used these values as training data to optimize the prediction method. The high prediction precision of this method was externally validated using an independent set of metabolites with a median relative error (MRE) of ~3%, better than conventional theoretical calculation. Using the SVR based prediction method, a large-scale predicted CCS database was generated for 35,203 metabolites in the Human Metabolome Database (HMDB). For each metabolite, five different ion adducts in positive and negative modes were predicted, accounting for 176,015 CCS values in total. Finally, improved metabolite identification accuracy was demonstrated using real biological samples. Conclusively, our results proved that the SVR based prediction method can accurately predict nitrogen CCS values (ΩN2) of metabolites from molecular descriptors, and effectively improve identification accuracy and efficiency in untargeted metabolomics. The predicted CCS database, namely, MetCCS, is freely available for non-commercial use.
Availability of MetCCS: http://www.zhulab.cn/MetCCS
Declaration: The MetCCS database is ONLY allowed for non-commercial use. Without a license, no one is allowed to copy, modify and distribute the MetCCS database. Since users can access MetCCS through our new web server, we will not provide downloadable version of the MetCCS database.
LipidCCS: Large-Scale and Precise Prediction of Collision Cross-Section Values for Lipids to Support Ion Mobility-Mass Spectrometry based Lipidomics
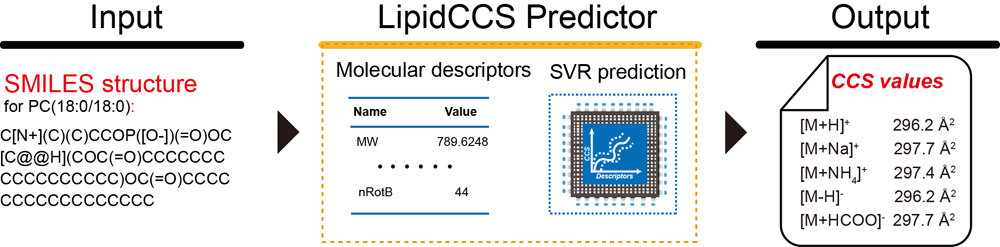
Brief introduction: Recently, ion mobility-mass spectrometry (IM-MS) showed great potential for lipidomics. However, the application of IM-MS to lipidomics is restrict by the limited number of available collision cross-section (CCS) values of lipids. In this work, we developed a new approach, namely, LipidCCS Predictor, to enable the large-scale and precise prediction of lipid CCS values, and to support the IM-MS based lipidomics. The newly developed LipidCCS is characterized by three innovative aspects: (1) it supports the prediction of lipid CCS values directly from SMILES structures; (2) a large set of lipid CCS values (458 in total) were experimentally measured to build the machine-learning algorithm based prediction model; (3) a new method to optimize the selection of molecular descriptors was developed to improve the prediction precision. Using independent external data sets, the prediction precision of LipidCCS was externally validated with a median relative error (MRE) of ~1% across different instruments (Agilent DTIM-MS and Waters TWIM-MS) and labs. A large-scale LipidCCS database containing 15,646 lipids and 63,434 CCS values was generated and demonstrated to effectively reduce false positive identifications of lipids in IM-MS based lipidomics workflow. Conclusively, our results proved that LipidCCS can precisely predict CCS values of lipids, and will be a valuable tool to support IM-MS based lipidomics.
Availability of LipidCCS: http://www.zhulab.cn/LipidCCS
Declaration: The LipidCCS database is ONLY allowed for non-commercial use. Without a license, no one is allowed to copy, modify and distribute the LipidCCS database.
Publications:
1. Z. Zhou, X. Shen, J. Tu, and Z.-J. Zhu* (Corresponding author), Large-Scale Prediction of Collision Cross-Section Values for Metabolites in Ion Mobility - Mass Spectrometry, Analytical Chemistry, 2016, 88, 1084-11091.
2. Z. Zhou, X. Xiong, and Z.-J. Zhu* (Corresponding author), MetCCS Predictor: A Web Server for Predicting Collision Cross-Section Values of Metabolite in Metabolomics, Bioinformatics, 2017, 33, 2235-2237.
3. Z. Zhou, J. Tu, X. Xiong, X. Shen, and Z.-J. Zhu* (Corresponding author), LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision to Support Ion Mobility-Mass Spectrometry based Lipidomics, Analytical Chemistry, 2017, 89, 9559–9566.
4. Z. Zhou†, J. Tu†, and Z.-J. Zhu*(Corresponding author), Advancing the Large-Scale CCS Database for Metabolomics and Lipidomics at the Machine-Learning Era, Current Opinion in Chemical Biology, 2018, 42, 34-41. (†, Co-first authors)
5. M. Luo, Z. Zhou, and Z.-J. Zhu* (Corresponding author), The Application of Ion Mobility-Mass Spectrometry in Untargeted Metabolomics: from Separation to Identification, Journal of Analysis and Testing, 2020, 4, 163–174.
6. T. Li, Y. Yin, Z. Zhou, J. Qiu, W. Liu, X. Zhang, K. He, Y. Cai, and Z.-J. Zhu*, Ion Mobility-based Sterolomics Reveals Spatially and Temporally Distinctive Sterol Lipids in the Mouse Brain, Nature Communications, 2021, 12: 4343. Web Link